TL;DR
Created a corpus of research articles related with COVID-19, automatically annotated with 10 biomedical entities of interest, namely Disorder, Species, Chemical or Drug, Gene or Protein, Enzyme, Anatomy, Biological Process, Molecular Function, Cellular Component, Pathway and microRNA.

The corpus is freely available and can be used to further research topics related with COVID-19, contributing to find insights towards a better understanding of the disease, in order to find effective drugs and reduce the pandemic impact.
Please follow the progress on Github, which already provides the CORD-19 corpus of full-text articles with more then 31 million biomedical annotations.
Download
Download the latest version of the COVID-19 annotated corpus.
Statistics
Overall corpus statistics:
- Number of abstracts: 17740
- Number of entity occurrences: 683349
- Number of unique entities: 29423
Number of annotations per entity type:
| Entity | # Occurrences | # Unique |
|---|---|---|
| Disorder | 183528 | 4477 |
| Species | 128356 | 2170 |
| Chemical or Drug | 70619 | 2768 |
| Gene and Protein | 51114 | 15025 |
| Enzyme | 7892 | 282 |
| Anatomy | 106401 | 2369 |
| Biological Process | 74286 | 1561 |
| Molecular Function | 15089 | 383 |
| Cellular Component | 39451 | 263 |
| Pathway | 6587 | 97 |
| microRNA | 26 | 28 |
Structure
Corpus file corpus/pubmed_YYYYMMDD.zip contains the following folders:
- json: article in JSON format from Pubmed;
- raw: article with text only;
- annotations: annotations in A1 format.
On each folder you can find one file per article, with the Pubmed ID on its name.
Articles
To collect articles related with COVID-19 from Pubmed, the following query was applied:
1
("2000"[Date - Publication] : "3000"[Date - Publication]) AND ((COVID-19) OR (Coronavirus) OR (Corona virus) OR (2019-nCoV) OR (SARS-CoV) OR (MERS-CoV) OR (Severe Acute Respiratory Syndrome) OR (Middle East Respiratory Syndrome) OR (2019 novel coronavirus disease[MeSH Terms]) OR (2019 novel coronavirus infection[MeSH Terms]) OR (2019-nCoV disease[MeSH Terms]) OR (2019-nCoV infection[MeSH Terms]) OR (coronavirus disease 2019[MeSH Terms]) OR (coronavirus disease-19[MeSH Terms]))
To collect the articles in JSON and then extract the text in raw format:
1
2
python scripts/pubmed/pubmed.py
python scripts/pubmed/json2raw.py
Please not that sentences in other languages than english are currently being discarded.
Resources
The following resources were applied to annotated each entity type:
- Disorder (DISO): UMLS
- Species (SPEC): NCBI Taxonomy
- Chemical or Drug (CHED): ChEBI
- Gene or Protein (PRGE): NER with CRFs and normalization with UniProt
- Enzyme (ENZY): ExPASy
- Anatomy (ANAT): Unified Medical Language System (UMLS)
- Biological Process (PROC): Gene Ontology (GO) and UMLS
- Molecular Function (FUNC): Gene Ontology (GO)
- Cellular Component (COMP): Gene Ontology (GO)
- Pathway (PATH): NCBI BioSystems
- microRNA (MRNA): miRBase
For more details please check the article. Unfortunately dictionaries could not be shared for download, due to UMLS usage license. Nevertheless, keep in mind that Disorder and Species entities were extended to include COVID-19 entities of interest.
Annotation
Neji is the tool used for NER (Named Entity Recognition) and normalization, which is optimized for biomedical scientific articles and provides an easy to use CLI. For more details please check the article.
The annotation script is available at scripts/pubmed/annotate.sh.
Visualization
brat is used to visualize the annotations in the articles. Find below the instructions to run the tool, create corpus for brat and visualize annotations.
Install and run brat
1
2
3
4
5
cd tools
unzip brat-1.3.zip
cd brat-1.3
./install.sh -u
python standalone.py
Create corpus for visualization
1
2
./scripts/pubmed/brat.sh
ln -s corpus/pubmed/brat tools/brat-1.3/data/covid19-corpus
Visualize corpus
Go to http://localhost:8001/index.xhtml#/covid19-corpus/ and wait for the articles to load:
 Figure: List of articles.
Figure: List of articles.
Double click in a document to visualize it:
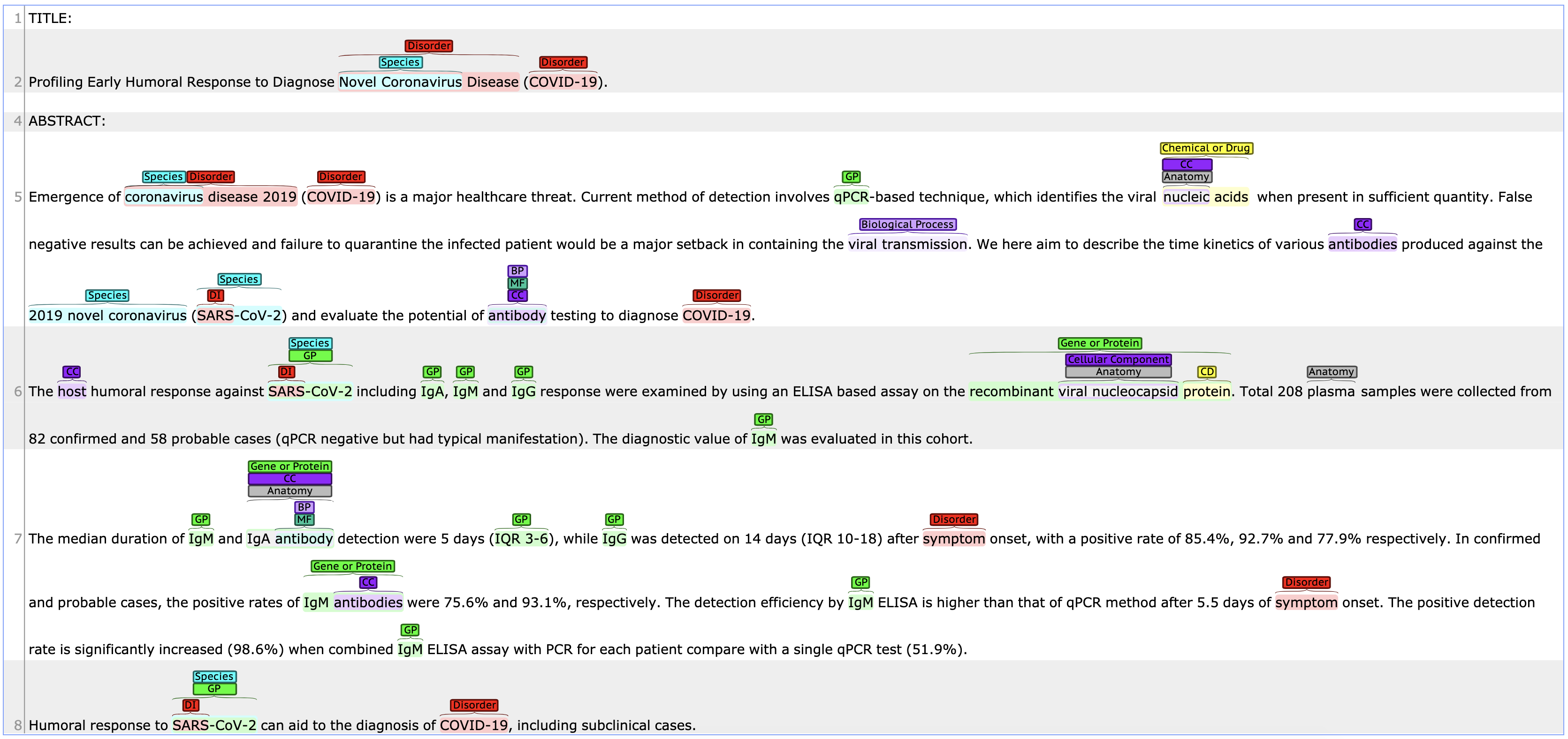 Figure: Article with annotations visualization.
Figure: Article with annotations visualization.
Example
Find below an example article in the provided document formats: JSON, Raw and A1.
JSON
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
{
"pubmed_id": "32198088",
"title": "Transmission potential and severity of COVID-19 in South Korea.",
"abstract": "Since the first case of 2019 novel coronavirus (COVID-19) identified on Jan 20, 2020 in South Korea, the number of cases rapidly increased, resulting in 6,284 cases including 42 deaths as of March 6, 2020. To examine the growth rate of the outbreak, we aimed to present the first study to report the reproduction number of COVID-19 in South Korea.\nThe daily confirmed cases of COVID-19 in South Korea were extracted from publicly available sources. By using the empirical reporting delay distribution and simulating the generalized growth model, we estimated the effective reproduction number based on the discretized probability distribution of the generation interval.\nWe identified four major clusters and estimated the reproduction number at 1.5 (95% CI: 1.4-1.6). In addition, the intrinsic growth rate was estimated at 0.6 (95% CI: 0.6, 0.7) and the scaling of growth parameter was estimated at 0.8 (95% CI: 0.7, 0.8), indicating sub-exponential growth dynamics of COVID-19. The crude case fatality rate is higher among males (1.1%) compared to females (0.4%) and increases with older age.\nOur results indicate early sustained transmission of COVID-19 in South Korea and support the implementation of social distancing measures to rapidly control the outbreak.",
"keywords": [
"COVID-19",
"Korea",
"coronavirus",
"reproduction number"
],
"journal": "International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases",
"publication_date": "2020-03-22",
"authors": [
{
"lastname": "Shim",
"firstname": "Eunha",
"initials": "E",
"affiliation": "Department of Mathematics, Soongsil University, 369 Sangdoro, Dongjak-Gu, Seoul, 06978 Republic of Korea. Electronic address: alicia@ssu.ac.kr."
},
{
"lastname": "Tariq",
"firstname": "Amna",
"initials": "A",
"affiliation": "Department of Population Health Sciences, School of Public Health, Georgia State University, Atlanta, GA, USA. Electronic address: atariq1@student.gsu.edu."
},
{
"lastname": "Choi",
"firstname": "Wongyeong",
"initials": "W",
"affiliation": "Department of Mathematics, Soongsil University, 369 Sangdoro, Dongjak-Gu, Seoul, 06978 Republic of Korea. Electronic address: chok10004@soongsil.ac.kr."
},
{
"lastname": "Lee",
"firstname": "Yiseul",
"initials": "Y",
"affiliation": "Department of Population Health Sciences, School of Public Health, Georgia State University, Atlanta, GA, USA. Electronic address: ylee97@student.gsu.edu."
},
{
"lastname": "Chowell",
"firstname": "Gerardo",
"initials": "G",
"affiliation": "Department of Population Health Sciences, School of Public Health, Georgia State University, Atlanta, GA, USA. Electronic address: gchowell@gsu.edu."
}
],
"methods": null,
"conclusions": null,
"results": "We identified four major clusters and estimated the reproduction number at 1.5 (95% CI: 1.4-1.6). In addition, the intrinsic growth rate was estimated at 0.6 (95% CI: 0.6, 0.7) and the scaling of growth parameter was estimated at 0.8 (95% CI: 0.7, 0.8), indicating sub-exponential growth dynamics of COVID-19. The crude case fatality rate is higher among males (1.1%) compared to females (0.4%) and increases with older age.",
"copyrights": "Copyright \u00a9 2020. Published by Elsevier Ltd.",
"doi": "10.1016/j.ijid.2020.03.031",
"xml": null
}
Raw
1
2
3
4
5
6
7
8
TITLE:
Transmission potential and severity of COVID-19 in South Korea.
ABSTRACT:
Since the first case of 2019 novel coronavirus (COVID-19) identified on Jan 20, 2020 in South Korea, the number of cases rapidly increased, resulting in 6,284 cases including 42 deaths as of March 6, 2020. To examine the growth rate of the outbreak, we aimed to present the first study to report the reproduction number of COVID-19 in South Korea.
The daily confirmed cases of COVID-19 in South Korea were extracted from publicly available sources. By using the empirical reporting delay distribution and simulating the generalized growth model, we estimated the effective reproduction number based on the discretized probability distribution of the generation interval.
We identified four major clusters and estimated the reproduction number at 1.5 (95% CI: 1.4-1.6). In addition, the intrinsic growth rate was estimated at 0.6 (95% CI: 0.6, 0.7) and the scaling of growth parameter was estimated at 0.8 (95% CI: 0.7, 0.8), indicating sub-exponential growth dynamics of COVID-19. The crude case fatality rate is higher among males (1.1%) compared to females (0.4%) and increases with older age.
Our results indicate early sustained transmission of COVID-19 in South Korea and support the implementation of social distancing measures to rapidly control the outbreak.
Annotations
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
T0 DISO 46 54 COVID-19
N0 Reference T0 UMLS:::DISO COVID-19
T1 SPEC 106 128 2019 novel coronavirus
N1 Reference T1 NCBI:2697049:T001:SPEC 2019 novel coronavirus
T2 DISO 130 138 COVID-19
N2 Reference T2 UMLS:::DISO COVID-19
T3 PROC 260 266 deaths
N3 Reference T3 GO:0016265::PROC deaths
T4 PROC 303 309 growth
N4 Reference T4 UMLS:C1621966:T042:PROC growth
N5 Reference T4 GO:0040007::PROC growth
T5 PROC 382 394 reproduction
N6 Reference T5 GO:0000003::PROC reproduction
T6 DISO 405 413 COVID-19
N7 Reference T6 UMLS:::DISO COVID-19
T7 DISO 459 467 COVID-19
N8 Reference T7 UMLS:::DISO COVID-19
T8 PROC 602 620 generalized growth
N9 Reference T8 GO:0040007::PROC generalized growth
T9 PROC 614 620 growth
N10 Reference T9 UMLS:C1621966:T042:PROC growth
T10 PROC 655 667 reproduction
N11 Reference T10 GO:0000003::PROC reproduction
T11 CHED 778 786 clusters
N12 Reference T11 CHEBI:33731:T103:CHED clusters
T12 PROC 805 817 reproduction
N13 Reference T12 GO:0000003::PROC reproduction
T13 PROC 878 884 growth
N14 Reference T13 UMLS:C1621966:T042:PROC growth
N15 Reference T13 GO:0040007::PROC growth
T14 PROC 949 955 growth
N16 Reference T14 UMLS:C1621966:T042:PROC growth
N17 Reference T14 GO:0040007::PROC growth
T15 PROC 1034 1040 growth
N18 Reference T15 UMLS:C1621966:T042:PROC growth
N19 Reference T15 GO:0040007::PROC growth
T16 DISO 1053 1061 COVID-19
N20 Reference T16 UMLS:::DISO COVID-19
T17 DISO 1231 1239 COVID-19
N21 Reference T17 UMLS:::DISO COVID-19
Next steps
Possible next steps to improve the COVID-19 corpus:
- Annotate “methods”, “results” and “conclusions” sections from JSON files;
- Further optimize resources to target entities related with COVID-19;
- Include additional entities of relevance;
- Annotate PMC and Elsevier full text articles;
- Collect co-occurrences to understand which entities might be related more often;
- Index articles and annotations and provide access to search tool.
Conclusion
I hope this annotated corpus helps to understand the COVID-19 disease better, towards finding better medication and to reduce the impact on society as much as possible. Please remember that your comments, suggestions and contributions are more than welcome.
Let’s kick the virus ass! ![]()

Comments